Proteomics
Four Core Genotype
Description here.
Heatmap of dataframe
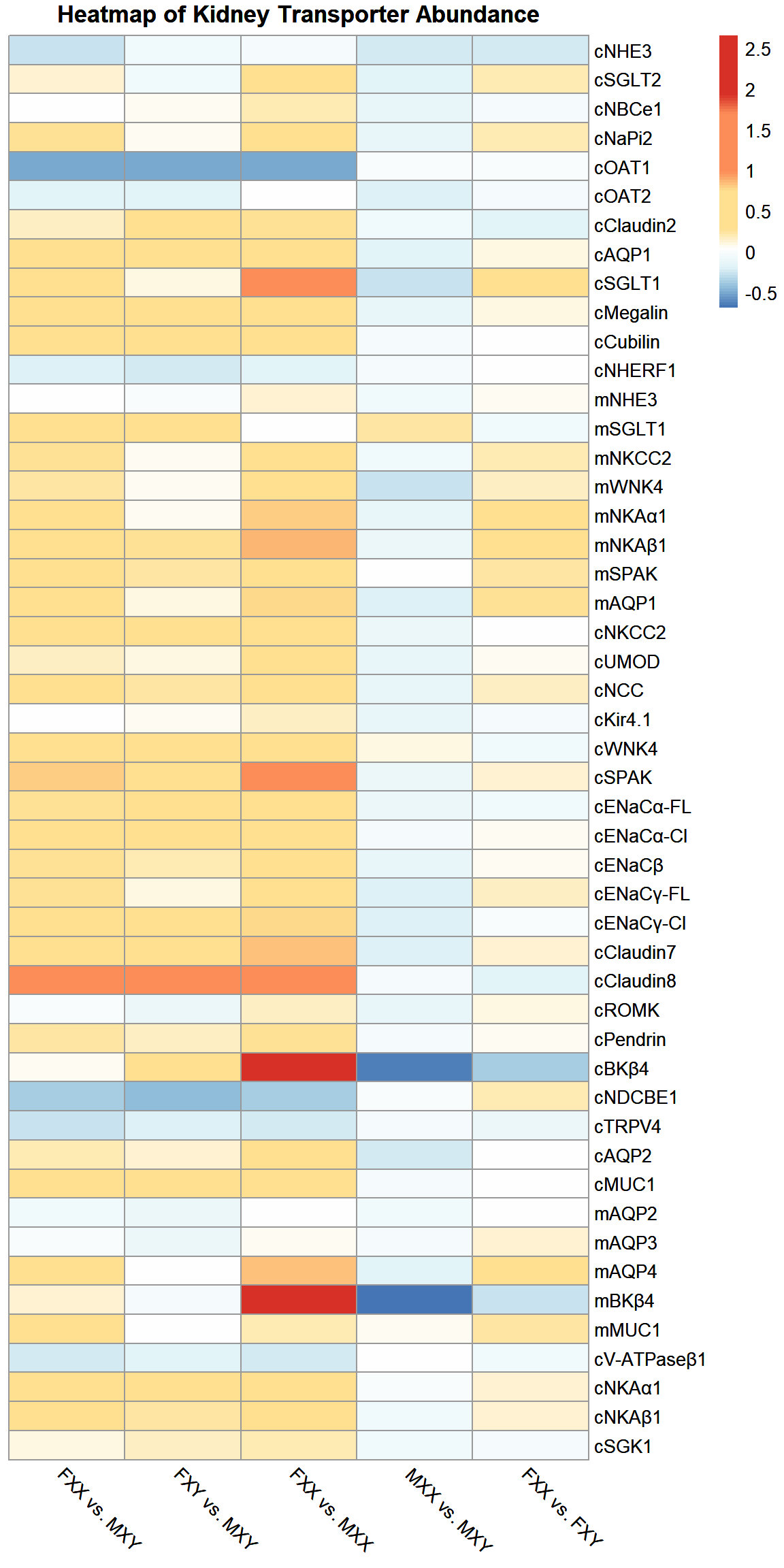
Table of dataframe
| FXX vs. MXY | FXY vs. MXY | FXX vs. MXX | MXX vs. MXY | FXX vs. FXY | |
|---|---|---|---|---|---|
| cNHE3 | -0.286700 | -0.0617 | -0.05167 | -0.245000 | -0.23670 |
| cSGLT2 | 0.113300 | -0.0733 | 0.34830 | -0.171700 | 0.20670 |
| cNBCe1 | 0.028330 | 0.0683 | 0.20670 | -0.146700 | -0.03833 |
| cNaPi2 | 0.255000 | 0.0533 | 0.46670 | -0.145000 | 0.19500 |
| cOAT1 | -0.510000 | -0.5070 | -0.52170 | -0.016670 | -0.01167 |
| cOAT2 | -0.183300 | -0.1630 | 0.03500 | -0.201700 | -0.02333 |
| cClaudin2 | 0.156700 | 0.4030 | 0.25830 | -0.070000 | -0.17500 |
| cAQP1 | 0.415000 | 0.2970 | 0.68170 | -0.156700 | 0.10330 |
| cSGLT1 | 0.745000 | 0.0917 | 1.41700 | -0.276700 | 0.60330 |
| cMegalin | 0.493300 | 0.3770 | 0.74330 | -0.135000 | 0.08667 |
| cCubilin | 0.555000 | 0.5170 | 0.61500 | -0.030000 | 0.04000 |
| cNHERF1 | -0.208300 | -0.2220 | -0.16500 | -0.051670 | 0.01667 |
| mNHE3 | 0.026670 | -0.0200 | 0.12170 | -0.076670 | 0.05167 |
| mSGLT1 | 0.330000 | 0.3530 | 0.02667 | 0.233300 | -0.06167 |
| mNKCC2 | 0.270000 | 0.0650 | 0.35670 | -0.060000 | 0.19170 |
| mWNK4 | 0.223300 | 0.0483 | 0.68670 | -0.255000 | 0.17000 |
| mNKAα1 | 0.548300 | 0.0633 | 0.80000 | -0.140000 | 0.45830 |
| mNKAβ1 | 0.690000 | 0.2770 | 0.87330 | -0.103300 | 0.32830 |
| mSPAK | 0.543300 | 0.2430 | 0.49670 | 0.030000 | 0.24000 |
| mAQP1 | 0.391700 | 0.0867 | 0.77500 | -0.215000 | 0.28000 |
| cNKCC2 | 0.416700 | 0.3600 | 0.56670 | -0.096670 | 0.04167 |
| cUMOD | 0.165000 | 0.1030 | 0.36830 | -0.145000 | 0.06000 |
| cNCC | 0.416700 | 0.2200 | 0.62000 | -0.126700 | 0.16000 |
| cKir4.1 | 0.020000 | 0.0667 | 0.17830 | -0.135000 | -0.04333 |
| cWNK4 | 0.495000 | 0.6200 | 0.37830 | 0.083330 | -0.07833 |
| cSPAK | 0.796700 | 0.6150 | 0.98670 | -0.095000 | 0.11500 |
| cENaCα-FL | 0.278300 | 0.3620 | 0.41670 | -0.096670 | -0.05833 |
| cENaCα-Cl | 0.493300 | 0.3970 | 0.55330 | -0.035000 | 0.07000 |
| cENaCβ | 0.256700 | 0.1850 | 0.46670 | -0.141700 | 0.06833 |
| cENaCγ-FL | 0.276700 | 0.0917 | 0.61830 | -0.206700 | 0.16670 |
| cENaCγ-Cl | 0.376700 | 0.3720 | 0.76330 | -0.215000 | 0.01000 |
| cClaudin7 | 0.461700 | 0.2980 | 0.82000 | -0.190000 | 0.13000 |
| cClaudin8 | 0.990000 | 1.4300 | 1.10500 | -0.048330 | -0.18000 |
| cROMK | -0.010000 | -0.0917 | 0.16500 | -0.150000 | 0.09000 |
| cPendrin | 0.220000 | 0.1480 | 0.26000 | -0.035000 | 0.06167 |
| cBKβ4 | 0.076670 | 0.7080 | 2.06300 | -0.643300 | -0.36500 |
| cNDCBE1 | -0.371700 | -0.4480 | -0.35670 | -0.001667 | 0.18830 |
| cTRPV4 | -0.276700 | -0.1880 | -0.23670 | -0.053330 | -0.10670 |
| cAQP2 | 0.193300 | 0.1450 | 0.53830 | -0.225000 | 0.04000 |
| cMUC1 | 0.621700 | 0.5680 | 0.70500 | -0.048330 | 0.03333 |
| mAQP2 | -0.061670 | -0.0917 | 0.02167 | -0.083330 | 0.03500 |
| mAQP3 | 0.001667 | -0.1120 | 0.04667 | -0.041670 | 0.12830 |
| mAQP4 | 0.505000 | 0.0433 | 0.83670 | -0.183300 | 0.47330 |
| mBKβ4 | 0.130000 | -0.0433 | 2.65700 | -0.690000 | -0.25830 |
| mMUC1 | 0.300000 | 0.0450 | 0.21000 | 0.075000 | 0.24170 |
| cV-ATPaseβ1 | -0.238300 | -0.1850 | -0.25000 | 0.023330 | -0.06333 |
| cNKAα1 | 0.525000 | 0.3330 | 0.54000 | -0.011670 | 0.14170 |
| cNKAβ1 | 0.365000 | 0.2300 | 0.48500 | -0.075000 | 0.12670 |
| cSGK1 | 0.111700 | 0.1720 | 0.20830 | -0.078330 | -0.05000 |
PT ACE2 KO
Description here.
Heatmap of dataframe
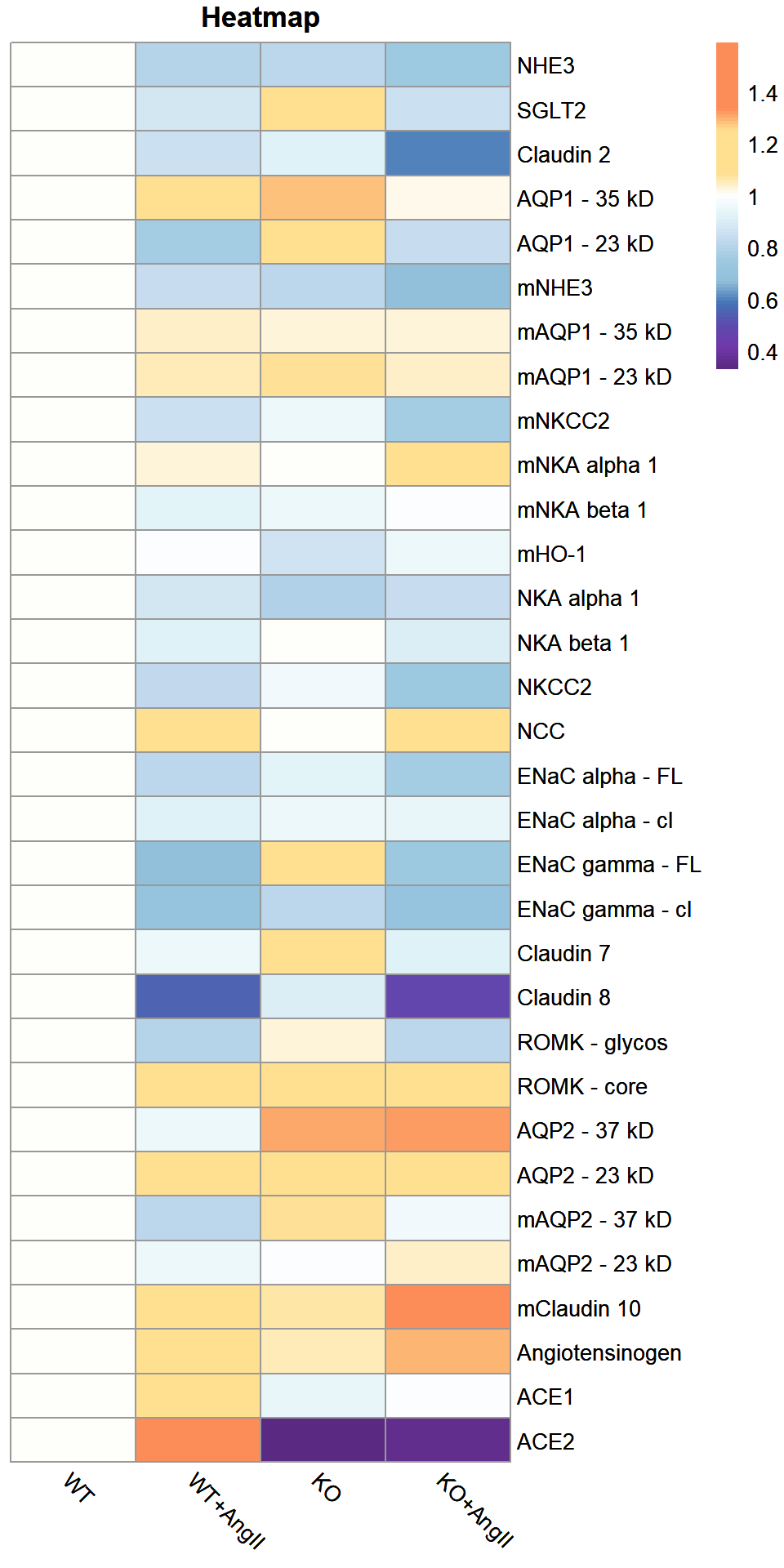
Table of dataframe
| WT | WT+AngII | KO | KO+AngII | |
|---|---|---|---|---|
| NHE3 | 1 | 0.80 | 0.82 | 0.75 |
| SGLT2 | 1 | 0.88 | 1.20 | 0.85 |
| Claudin 2 | 1 | 0.85 | 0.91 | 0.60 |
| AQP1 - 35 kD | 1 | 1.10 | 1.28 | 1.02 |
| AQP1 - 23 kD | 1 | 0.76 | 1.23 | 0.84 |
| mNHE3 | 1 | 0.84 | 0.82 | 0.68 |
| mAQP1 - 35 kD | 1 | 1.04 | 1.03 | 1.03 |
| mAQP1 - 23 kD | 1 | 1.05 | 1.08 | 1.04 |
| mNKCC2 | 1 | 0.85 | 0.95 | 0.76 |
| mNKA alpha 1 | 1 | 1.03 | 1.01 | 1.09 |
| mNKA beta 1 | 1 | 0.93 | 0.95 | 0.99 |
| mHO-1 | 1 | 0.99 | 0.86 | 0.96 |
| NKA alpha 1 | 1 | 0.88 | 0.79 | 0.84 |
| NKA beta 1 | 1 | 0.92 | 1.00 | 0.90 |
| NKCC2 | 1 | 0.83 | 0.97 | 0.74 |
| NCC | 1 | 1.20 | 1.01 | 1.10 |
| ENaC alpha - FL | 1 | 0.82 | 0.93 | 0.77 |
| ENaC alpha - cl | 1 | 0.92 | 0.96 | 0.94 |
| ENaC gamma - FL | 1 | 0.68 | 1.10 | 0.74 |
| ENaC gamma - cl | 1 | 0.70 | 0.81 | 0.70 |
| Claudin 7 | 1 | 0.95 | 1.09 | 0.92 |
| Claudin 8 | 1 | 0.55 | 0.90 | 0.47 |
| ROMK - glycos | 1 | 0.80 | 1.03 | 0.82 |
| ROMK - core | 1 | 1.10 | 1.10 | 1.23 |
| AQP2 - 37 kD | 1 | 0.96 | 1.31 | 1.32 |
| AQP2 - 23 kD | 1 | 1.15 | 1.22 | 1.13 |
| mAQP2 - 37 kD | 1 | 0.82 | 1.08 | 0.97 |
| mAQP2 - 23 kD | 1 | 0.96 | 0.99 | 1.04 |
| mClaudin 10 | 1 | 1.22 | 1.07 | 1.51 |
| Angiotensinogen | 1 | 1.13 | 1.05 | 1.30 |
| ACE1 | 1 | 1.12 | 0.94 | 0.99 |
| ACE2 | 1 | 1.59 | 0.33 | 0.36 |